1
2
3
4
5
6
7
8
9
10
11
12
| /MLOps-Breast-Cancer/Kafka$ curl -X POST http://localhost:8083/connectors -H "Content-Type: application/json" -d @source_connector.json
{"name":"postgres-source-connector","config":{"connector.class":"io.confluent.connect.jdbc.JdbcSourceConnector","connection.url":"jdbc:postgresql://postgres-server:5432/Breast_Cancer","connection.user":"zerohertz","connection.password":"qwer123!","table.whitelist":"breast_cancer_data","topic.prefix":"postgres-source-","topic.creation.default.partitions":"1","topic.creation.default.replication.factor":"1","mode":"incrementing","incrementing.column.name":"id","tasks.max":"2","transforms":"TimestampConverter","transforms.TimestampConverter.type":"org.apache.kafka.connect.transforms.TimestampConverter$Value","transforms.TimestampConverter.field":"timestamp","transforms.TimestampConverter.format":"yyyy-MM-dd HH:mm:ss.S","transforms.TimestampConverter.target.type":"string","name":"postgres-source-connector"},"tasks":[],"type":"source"}%
/MLOps-Breast-Cancer/Kafka$ curl -X GET http://localhost:8083/connectors
["postgres-source-connector"]%
/MLOps-Breast-Cancer/Kafka$ curl -X GET http://localhost:8083/connectors/postgres-source-connector
{"name":"postgres-source-connector","config":{"connector.class":"io.confluent.connect.jdbc.JdbcSourceConnector","transforms.TimestampConverter.target.type":"string","incrementing.column.name":"id","topic.creation.default.partitions":"1","connection.password":"qwer123!","transforms.TimestampConverter.field":"timestamp","tasks.max":"2","transforms.TimestampConverter.type":"org.apache.kafka.connect.transforms.TimestampConverter$Value","transforms":"TimestampConverter","transforms.TimestampConverter.format":"yyyy-MM-dd HH:mm:ss.S","table.whitelist":"breast_cancer_data","mode":"incrementing","topic.prefix":"postgres-source-","connection.user":"zerohertz","topic.creation.default.replication.factor":"1","name":"postgres-source-connector","connection.url":"jdbc:postgresql://postgres-server:5432/Breast_Cancer"},"tasks":[{"connector":"postgres-source-connector","task":0}],"type":"source"}%
$ kcat -L -b localhost:9092
...
topic "postgres-source-breast_cancer_data" with 1 partitions:
partition 0, leader 1, replicas: 1, isrs: 1
...
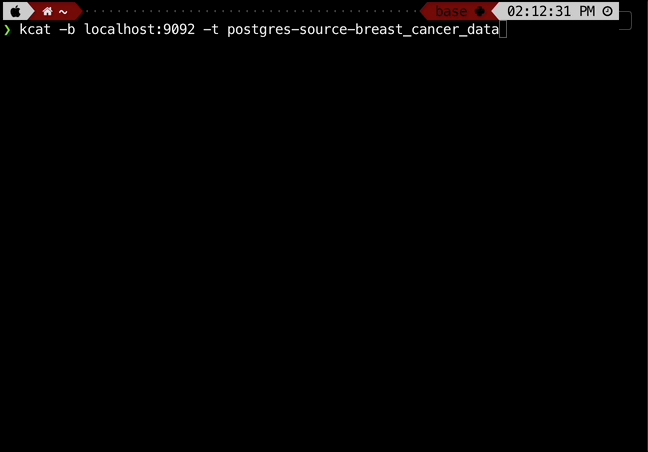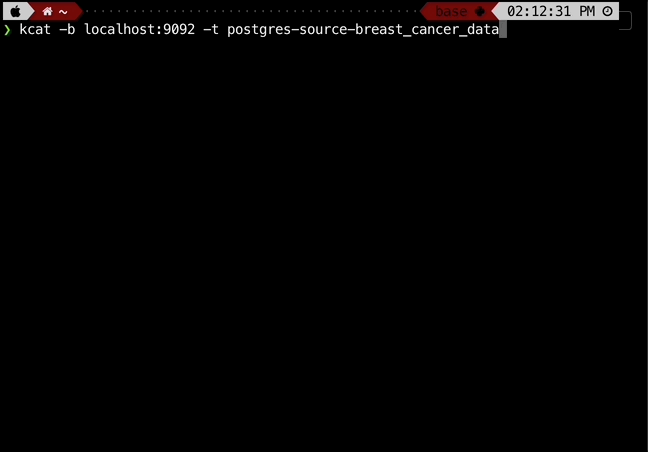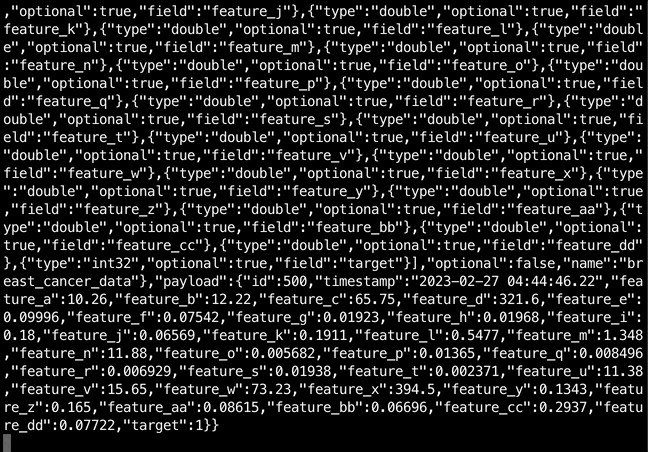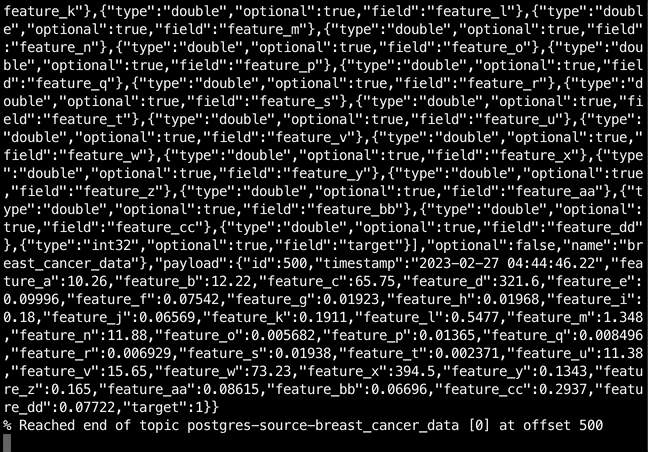
$ kcat -b localhost:9092 -t postgres-source-breast_cancer_data
|